3. Optimization#
This exercise is devoted to:
convex and non-convex optimization,
how to use different optimizer with PyTorch, and
how to apply some common tricks of optimization
Imports and Utils
import matplotlib
import matplotlib.pyplot as plt
import numpy as np
import torch
from torch import nn
def plot_progress(func, track, losses, func_min, torch=False):
"""
Adapted from
https://colab.research.google.com/github/davidbau/how-to-read-pytorch/blob/master/notebooks/3-Pytorch-Optimizers.ipynb
"""
# Draw the contours of the objective function, and x, and y
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 5))
xs = np.linspace(-2.5, 2.5, 100)
ys = np.linspace(-1, 4, 100)
X, Y = np.meshgrid(xs, ys)
Z = func(np.concatenate([np.expand_dims(X, 0), np.expand_dims(Y, 0)]))
Z = np.array(Z)
levels = np.linspace(Z.min(), Z.max(), 100)
manual_levels = np.array([5, 20, 100])
levels = np.concatenate(((Z.min()+0.001) * manual_levels, levels))
levels.sort()
ax1.contour(X, Y, Z, levels=levels, cmap="bwr", alpha=0.5)
track = np.stack(track).T # (2,n)
for i in range(len(losses)-1):
ax1.plot(track[0, i:i+2], track[1, i:i+2], marker='o',
color='k', alpha=0.2 + i/(len(losses)-1) * 0.3)
ax1.scatter(func_min[0], func_min[1], s=80, c='g', marker=(5, 1))
ax1.set_title('progress of x')
ax1.set_ylim(-1.0, 4)
ax1.set_xlim(-2.5, 2.5)
ax1.set_ylabel('x_2')
ax1.set_xlabel('x_1')
ax2.set_title('progress of loss')
ax2.xaxis.set_major_locator(matplotlib.ticker.MaxNLocator(integer=True))
for i in range(len(losses)-1):
ax2.plot([i, i+1], losses[i:i+2], marker='o',
color='k', alpha=0.2 + i/(len(losses)-1) * 0.3)
ax2.hlines(func(func_min), 0, len(losses), colors='g')
# ax2.plot(range(len(losses)), losses, marker='o', color='k')
ax2.set_ylabel('objective')
ax2.set_xlabel('iteration')
fig.show()
3.1. With Analytical Derivatives#
3.1.1. Case Study: Convex vs Non-Convex Optimization#
In the convex optimization case, we look at the Rosenbrock function
\(f_1(x) = (x_2-x_1^2)^2 + 0.01 \cdot (1-x_1)^2\)
The non-convex case emerges if we add a sinusoidal signal in \(x\), i.e.
\(f_2(x) = (x_2-x_1^2)^2 + 0.01 \cdot (1-x_1)^2 + 0.1 \cdot (1 + \cos(x_1))\)
def f1(x, use_torch=False):
"""Rosenbrock function
Has one local and global minimum at f(1,1)=0"""
return (x[1]-x[0] ** 2) ** 2 + 0.01 * (1-x[0]) ** 2
def grad_f1(x):
"""Gradient of Rosenbrock function"""
dx0 = - 2 * (x[1] - x[0] ** 2) * 2 * x[0] - 2 * 0.01 * (1-x[0])
dx1 = 2 * (x[1] - x[0] ** 2)
return np.array([dx0, dx1])
def hess_f1(x):
"""Hessian of Rosenbrock function"""
dxdx = 8*x[0]**2 - 4*(x[1]-x[0]**2) + 2*0.01
dxdy = dydx = -4*x[0]
dydy = 2
return np.array([[dxdx, dxdy], [dydx, dydy]])
f1_min = np.array([1.0, 1.0])
def f2(x, use_torch=False):
"""Extended Rosenbrock function
Has one global minimum f(0.63, 0.40)=0.00137 and many local minima"""
regular_rosenbrock = (x[1]-x[0] ** 2) ** 2 + 0.01 * (1-x[0]) ** 2
if use_torch:
return regular_rosenbrock + 0.1 * (1 + torch.cos(5*x[0]))
else:
return regular_rosenbrock + 0.1 * (1 + np.cos(5*x[0]))
def grad_f2(x):
"""Gradient of extended Rosenbrock function"""
dx0 = - 2 * (x[1] - x[0] ** 2) * 2 * x[0] - 2 * \
0.01 * (1-x[0]) - 0.1 * np.sin(5*x[0]) * 5
dx1 = 2 * (x[1] - x[0] ** 2)
return np.array([dx0, dx1])
def hess_f2(x):
"""Hessian of extended Rosenbrock function"""
dxdx = 8*x[0]**2 - 4*(x[1]-x[0]**2) + 2*0.01 - 0.1 * np.cos(5*x[0]) * 5 * 5
dxdy = dydx = -4*x[0]
dydy = 2
return np.array([[dxdx, dxdy], [dydx, dydy]])
f2_min = np.array([0.63, 0.40])
selected_function = 1
if selected_function == 1:
func = f1
grad_func = grad_f1
hess_func = hess_f1
func_min = f1_min
else:
func = f2
grad_func = grad_f2
hess_func = hess_f2
func_min = f2_min
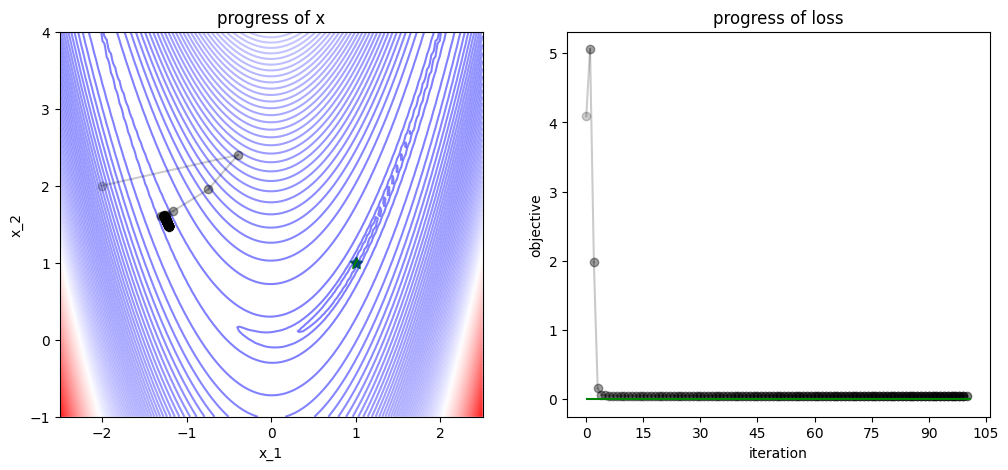
3.1.2. Gradient Descent (revised)#
num_iters = 100
x = np.array([-2., 2])
track, losses = [x], [func(x)]
def gd_step(x, grad, lr=0.1):
# GD step
x = x - lr * grad
return x
for iter in range(num_iters):
# apply gradient descent
grad = grad_func(x)
x = gd_step(x=x, grad=grad)
# logging
track.append(x)
losses.append(func(x))
plot_progress(func, track, losses, func_min)
/var/folders/7g/3mxmtrb16h7gh3hsxzbkh_5w0000gn/T/ipykernel_19923/4038594132.py:52: UserWarning: FigureCanvasAgg is non-interactive, and thus cannot be shown
fig.show()
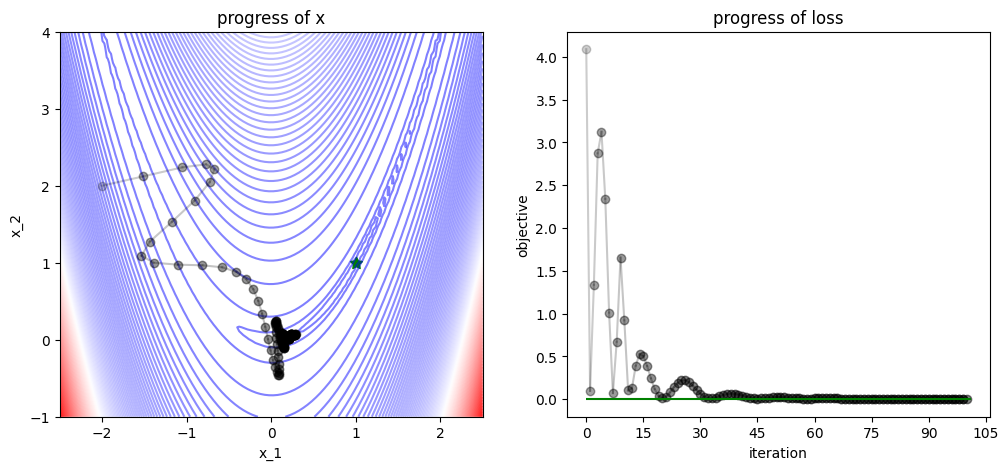
3.1.3. Gradient Descent with Momentum#
num_iters = 100
x = np.array([-2., 2])
v = np.zeros(x.shape)
track, losses = [x], [func(x)]
def gd_with_momentum_step(x, v, grad, lr=0.03, beta=0.9):
# GD with momentum step
v = beta*v + grad
x = x - lr * v
return (x, v)
for iter in range(num_iters):
# apply gradient descent with momentum
grad = grad_func(x)
x, v = gd_with_momentum_step(x=x, v=v, grad=grad)
# logging
track.append(x)
losses.append(func(x))
plot_progress(func, track, losses, func_min)
/var/folders/7g/3mxmtrb16h7gh3hsxzbkh_5w0000gn/T/ipykernel_19923/4038594132.py:52: UserWarning: FigureCanvasAgg is non-interactive, and thus cannot be shown
fig.show()
3.1.4. Adam#
num_iters = 100
x = np.array([-2., 2])
v = np.zeros(x.shape)
s = np.zeros(x.shape)
track, losses = [x], [func(x)]
def adam_step(x, v, s, grad, t, lr=.9, beta1=0.9, beta2=0.999):
# Adam step
v = beta1 * v + (1 - beta1) * grad
s = beta2 * s + (1 - beta2) * grad ** 2
v_hat = v / (1 - beta1 ** t)
s_hat = s / (1 - beta2 ** t)
# the small number in the next line is added to avoid divisions by zero
x = x - lr * v_hat / (np.sqrt(s_hat) + 0.0000001)
return (x, v, s)
for iter in range(num_iters):
# apply Adam
grad = grad_func(x)
x, v, s = adam_step(x=x, v=v, s=s, grad=grad, t=(iter+1))
# logging
track.append(x)
losses.append(func(x))
plot_progress(func, track, losses, func_min)
/var/folders/7g/3mxmtrb16h7gh3hsxzbkh_5w0000gn/T/ipykernel_19923/4038594132.py:52: UserWarning: FigureCanvasAgg is non-interactive, and thus cannot be shown
fig.show()
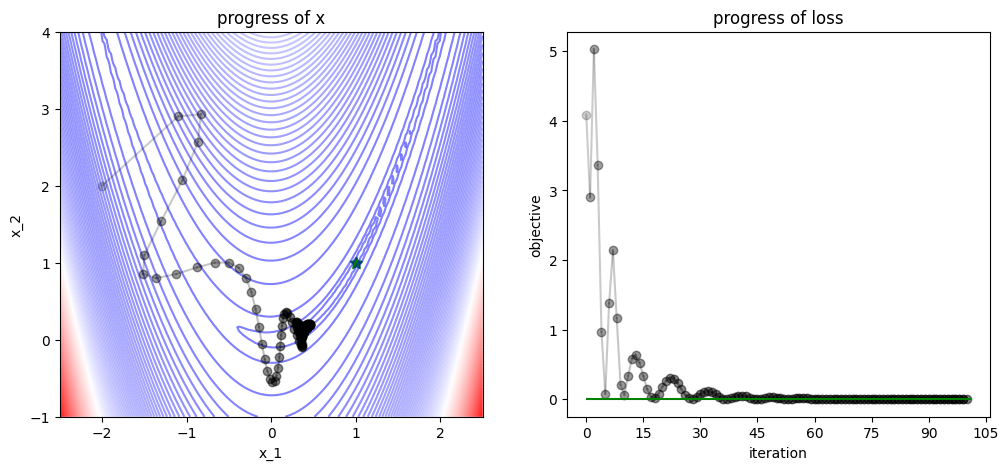
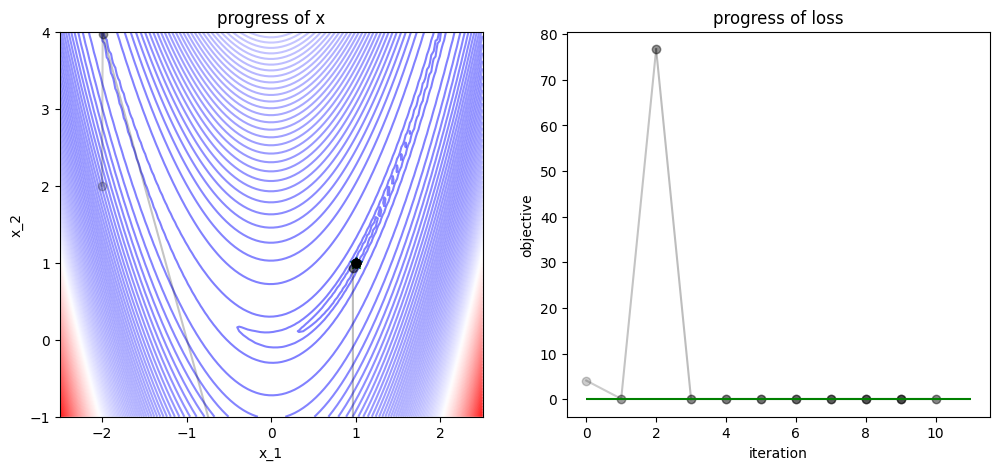
3.1.5. Newton Optimizer#
num_iters = 10
x = np.array([-2., 2])
track, losses = [x], [func(x)]
def newton_step(x, grad, hess, lr=1.0):
# Newton's method step
hess_inv = np.linalg.inv(hess)
x = x - lr * hess_inv @ grad
# equivalent to `x = x - lr * np.dot(hess_inv, grad)`
return x
for iter in range(num_iters):
# apply Newton's method
grad = grad_func(x)
hess = hess_func(x)
x = newton_step(x=x, grad=grad, hess=hess)
# logging
track.append(x)
losses.append(func(x))
plot_progress(func, track, losses, func_min)
/var/folders/7g/3mxmtrb16h7gh3hsxzbkh_5w0000gn/T/ipykernel_19923/4038594132.py:52: UserWarning: FigureCanvasAgg is non-interactive, and thus cannot be shown
fig.show()
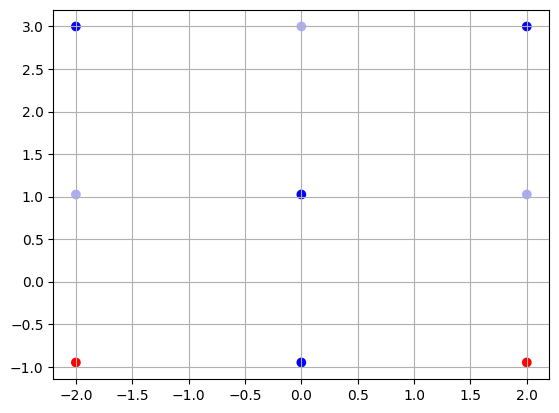
3.1.6. Derivative-Free Optimization#
Here, we compare two seemingly very similar algorithms: Grid Search and Random Search.
We assume that we know the range of the parameters in which the minimum is contained. We then just explore the region either in a structered way (grid search) or by random sampling (random search).
samples_per_dim = 3
# grid search
xs = np.linspace(-2.0, 2.0, samples_per_dim)
ys = np.linspace(-0.95, 3.0, samples_per_dim)
X, Y = np.meshgrid(xs, ys)
Z = np.zeros(X.shape)
for i in range(len(xs)):
for j in range(len(ys)):
Z[i, j] = func(np.array([X[i, j], Y[i, j]]))
plt.scatter(X.flatten(), Y.flatten(), c=Z.flatten(), cmap='bwr')
plt.grid()
idx = Z == Z.min()
print(
f"Our best guess is x={X[idx]}, y={Y[idx]}, and loss = {func(np.array([X[idx], Y[idx]]))}")
Our best guess is x=[0.], y=[-0.95], and loss = [0.9125]
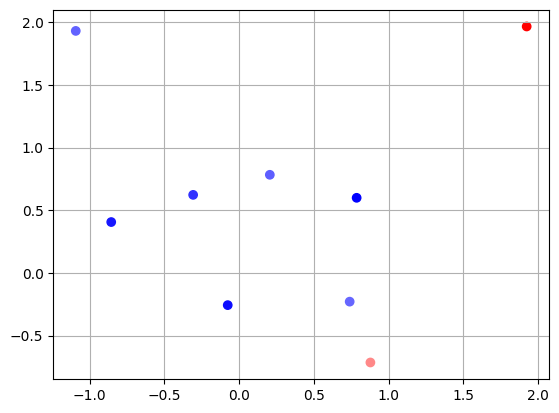
# random search
np.random.seed(123)
# sample randomly samples_per_dim**2 points on the x and y axis
X = np.random.uniform(-2.0, 2.0, int(samples_per_dim**2))
Y = np.random.uniform(-0.95, 3.0, int(samples_per_dim**2))
# evaluate loss at each of these points
Z = np.zeros(X.shape)
for i in range(len(Z)):
Z[i] = func(np.array([X[i], Y[i]]))
plt.scatter(X, Y, c=Z, cmap='bwr')
plt.grid()
idx = Z == Z.min()
print(
f"Our best guess is x={X[idx]}, y={Y[idx]}, and loss = {func(np.array([X[idx], Y[idx]]))}")
Our best guess is x=[0.78587674], y=[0.5988642], and loss = [0.0008096]
3.2. With PyTorch#
3.2.1. Gradient Descent#
num_iters = 100
x_init = torch.tensor([-2.0, 2.0])
x = x_init.clone()
x.requires_grad = True
optimizer = torch.optim.SGD([x], lr=0.1)
track = [x.detach().clone().numpy()]
losses = [func(x, use_torch=True).detach().numpy()]
for iter in range(num_iters):
loss = func(x, use_torch=True)
optimizer.zero_grad()
loss.backward()
optimizer.step()
track.append(x.detach().clone().numpy())
losses.append(loss.detach().numpy())
plot_progress(func, track, losses, func_min)
/var/folders/7g/3mxmtrb16h7gh3hsxzbkh_5w0000gn/T/ipykernel_19923/4038594132.py:52: UserWarning: FigureCanvasAgg is non-interactive, and thus cannot be shown
fig.show()
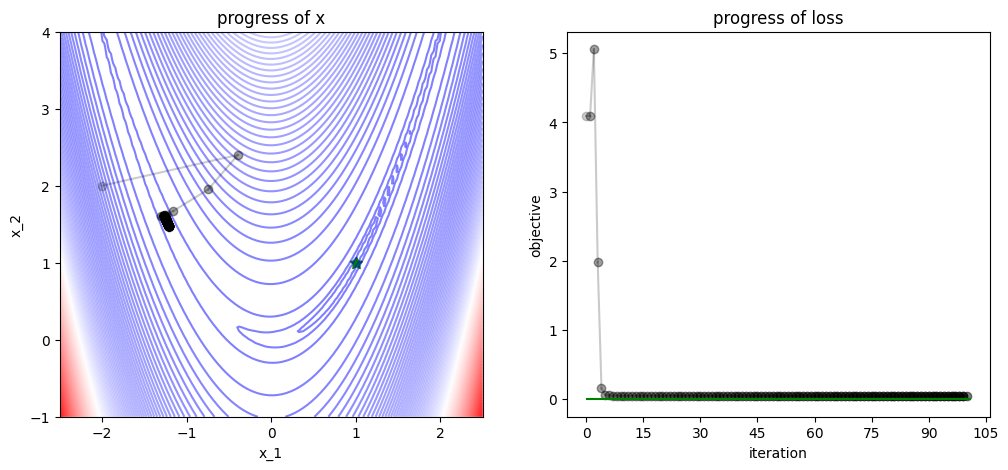
3.2.2. Adam#
num_iters = 100
x_init = torch.tensor([-2.0, 2.0])
x = x_init.clone()
x.requires_grad = True
optimizer = torch.optim.Adam([x], lr=1.0, betas=(0.9, 0.95))
track = [x.detach().clone().numpy()]
losses = [func(x, use_torch=True).detach().numpy()]
for iter in range(num_iters):
loss = func(x, use_torch=True)
optimizer.zero_grad()
loss.backward()
optimizer.step()
track.append(x.detach().clone().numpy())
losses.append(loss.detach().numpy())
plot_progress(func, track, losses, func_min)
/var/folders/7g/3mxmtrb16h7gh3hsxzbkh_5w0000gn/T/ipykernel_19923/4038594132.py:52: UserWarning: FigureCanvasAgg is non-interactive, and thus cannot be shown
fig.show()
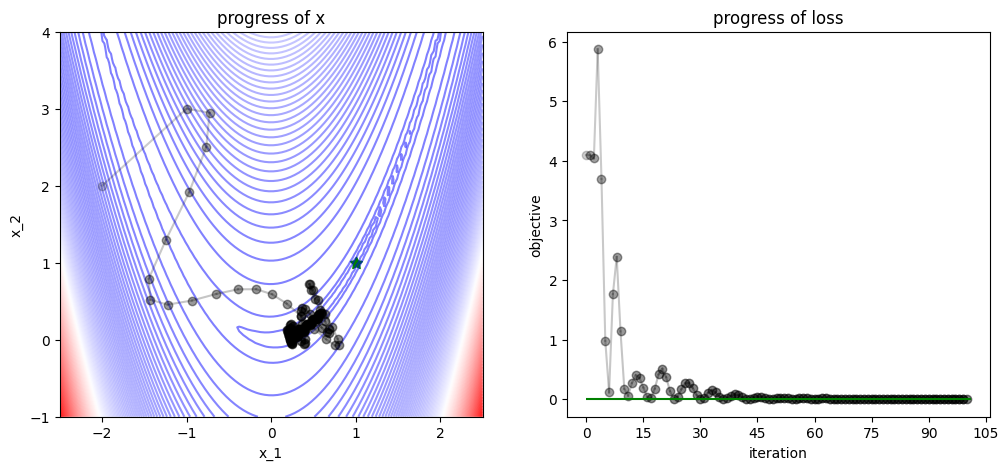
3.2.3. L-BFGS#
num_iters = 100
x_init = torch.tensor([-2.0, 2.0])
x = x_init.clone()
x.requires_grad = True
optimizer = torch.optim.LBFGS([x], lr=1.0)
track = [x.detach().clone().numpy()]
losses = [func(x, use_torch=True).detach().numpy()]
for iter in range(num_iters):
# LBFGS requres a `closure` for the approximation of the hessian
# typically it is the gradient:
def closure():
"""
from the [documentation](https://pytorch.org/docs/stable/generated/torch.optim.LBFGS.html):
"A closure that reevaluates the model and returns the loss."
More information on the closure here:
https://pytorch.org/docs/stable/optim.html
"""
optimizer.zero_grad()
loss = func(x, use_torch=True)
loss.backward()
return loss
optimizer.step(closure)
track.append(x.detach().clone().numpy())
losses.append(func(x, use_torch=True).detach().numpy())
optimizer.step(closure)
plot_progress(func, track, losses, func_min)
/var/folders/7g/3mxmtrb16h7gh3hsxzbkh_5w0000gn/T/ipykernel_19923/4038594132.py:52: UserWarning: FigureCanvasAgg is non-interactive, and thus cannot be shown
fig.show()
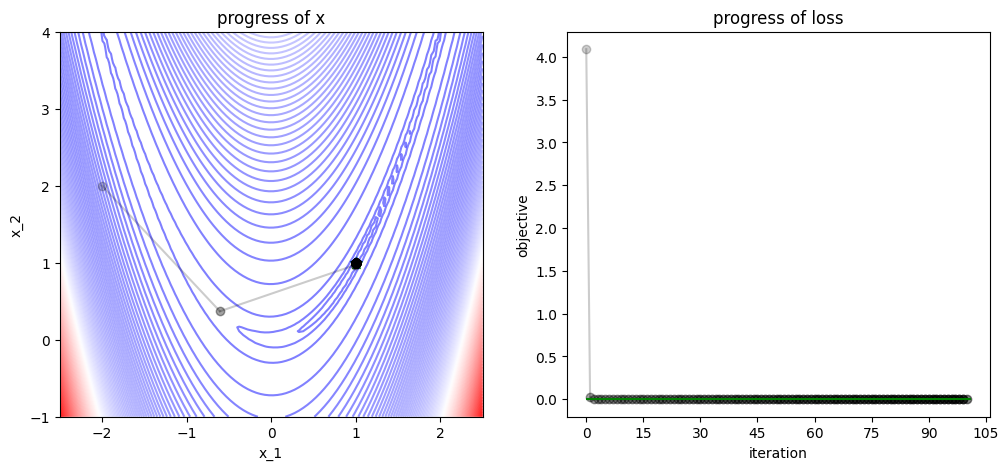
3.3. Tricks of Optimization#
# plot util
def plot_progress_polynomial(losses, model, x_data, y_data):
# visualization
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 5))
# left plot
x_linspace = torch.linspace(-1.5, 1.5, 31).unsqueeze(1)
with torch.no_grad():
y_pred_np = model(x_linspace).data.squeeze().numpy()
# for plotting we transfer everything from torch to numpy
x_data_np = x_data.detach().squeeze().numpy()
y_data_np = y_data.detach().squeeze().numpy()
x_np = x_linspace.squeeze().numpy()
# plot performance
ax1.plot(x_data_np, y_data_np, 'go', label='True data')
ax1.plot(x_np, y_pred_np, '--', label='Predictions')
ax1.set_ylim([-3, 3])
ax1.legend()
ax1.grid()
# right plot
ax2.plot(losses)
ax2.set_xlabel('iteration')
ax2.set_ylabel('loss')
ax2.grid()
plt.tight_layout()
plt.show()
# train util
def train(num_iters, model, optimizer, criterion, x_data, y_data, losses):
print("## Parameters before training")
for name, param in model.named_parameters():
print(name, ": ", param.data)
for iter in range(num_iters):
def closure():
optimizer.zero_grad()
y_pred = model(x_data)
loss = criterion(y_pred, y_data)
loss.backward()
return loss
optimizer.step(closure)
losses.append(closure().detach().item())
# Caution: LBFGS converges very fast, but will not always converge to the
# true minimum. Check the left plot below to verify validity.
print("## Parameters after training:")
for name, param in model.named_parameters():
print(name, ": ", param.data)
print(f"## Loss at the end of training = {losses[-1]}")
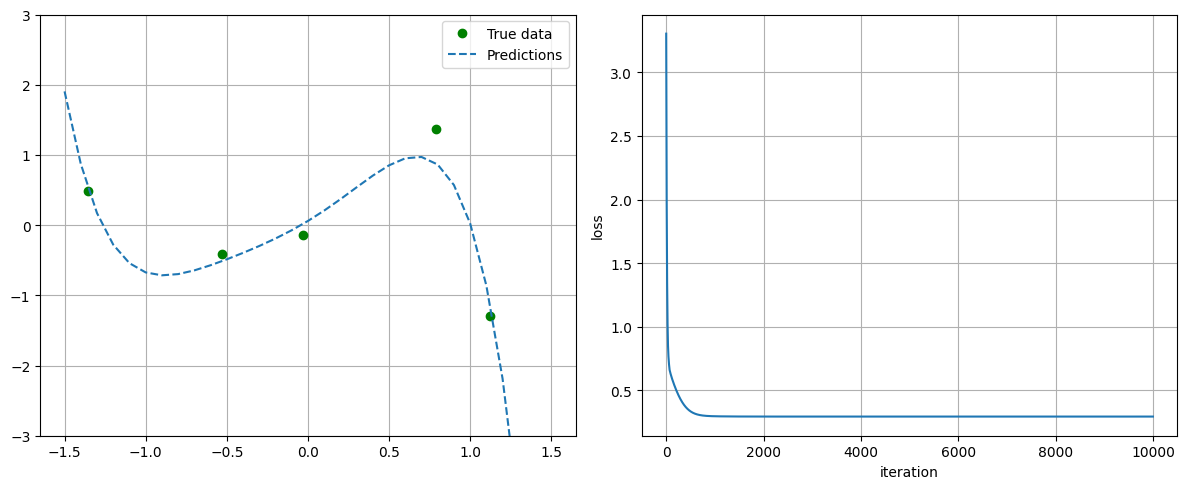
3.3.1. Case Study: Polynomial Linear Regression#
Note: we move back to the typical ML notation with measurement pairs \(\left\{x^{(i)}, y^{\text {(i)}}\right\}_{i=1,...m}\), model \(h(x)\) which approximates \(y\), and loss \(\mathcal{L}\)
In this specific case we look at polynomial linear regression, i.e.
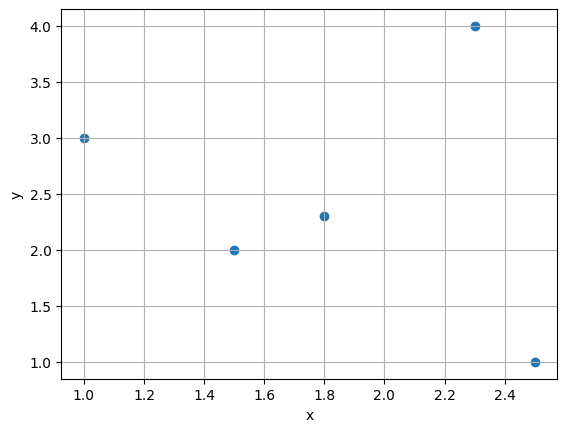
We work with a very simple artificial dataset consisting of 5 measurement pairs given below.
x_data = torch.tensor([[1.0, 1.5, 1.8, 2.3, 2.5]]).T
y_data = torch.tensor([[3.0, 2., 2.3, 4., 1.]]).T
print(x_data.shape, y_data.shape)
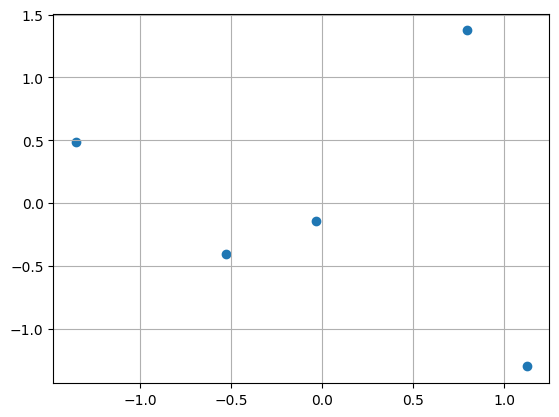
plt.scatter(x_data, y_data); plt.grid()
plt.xlabel('x'); plt.ylabel('y'); plt.show()
torch.Size([5, 1]) torch.Size([5, 1])
3.3.1.1. Input/Output Normalization#
Trick Nr. 1: Input/Output normalization to zero-centered and std=1 variables.
# TODO: vital for an efficient training process
x_mean = x_data.mean()
x_std = x_data.std()
x_data = (x_data-x_mean)/x_std
y_mean = y_data.mean()
y_std = y_data.std()
y_data = (y_data-y_mean)/y_std
plt.scatter(x_data, y_data); plt.grid()
3.3.2. Choice of Model Complexity#
Let’s first construct a linear model and set up an optimization with Adam.
# model
class PolynomialModel(nn.Module):
""" from https://soham.dev/posts/polynomial-regression-pytorch"""
def __init__(self, degree):
super().__init__()
self._degree = degree
# 1 is the dimension of the output y
self.linear = nn.Linear(self._degree, 1)
def forward(self, x):
return self.linear(self._polynomial_features(x))
def _polynomial_features(self, x):
return torch.cat([x ** i for i in range(1, self._degree + 1)], 1)
# x.shape = [B,F] where B is the batch size and F is the number of features
Exercise
Try various values for the degree of the polynomial. When does overfitting begin?
Keep in mind: polynomial with degree
dis able to perfectly fitd+1points
# TODO: try our different polynomial degrees
polynomial_degree = 3
model = PolynomialModel(polynomial_degree)
# Use LBFGS for fast convergence, but check validity of solution.
# Remember: LBFGS is only an approximate 2nd order optimizer
# optimizer = torch.optim.LBFGS(model.parameters())
optimizer = torch.optim.Adam(model.parameters(), lr=0.1)
criterion = nn.MSELoss()
losses = []
num_iters = 100
train(num_iters, model, optimizer, criterion, x_data, y_data, losses)
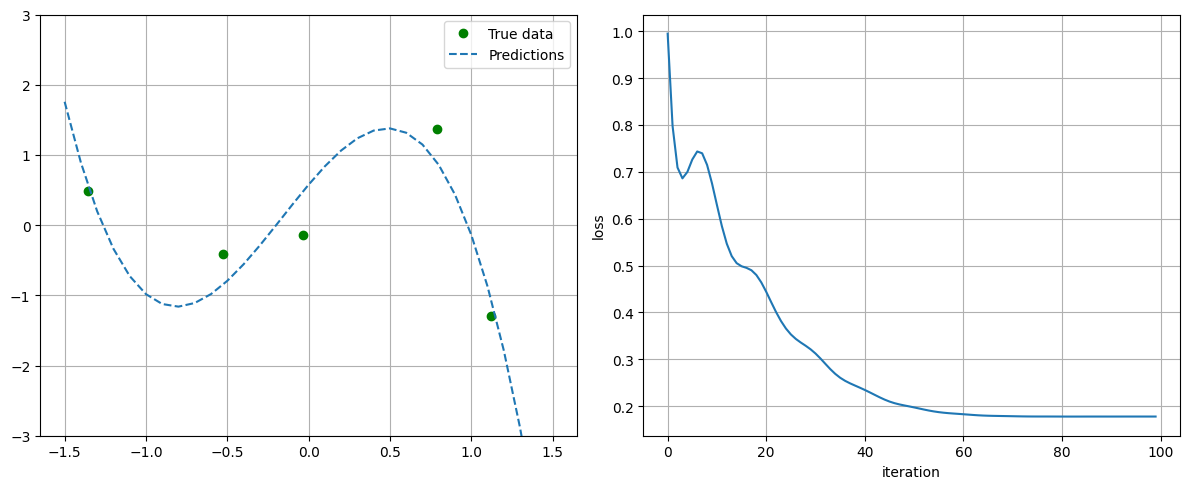
plot_progress_polynomial(losses, model, x_data, y_data)
## Parameters before training
linear.weight : tensor([[-0.2872, -0.3925, -0.4374]])
linear.bias : tensor([-0.4289])
## Parameters after training:
linear.weight : tensor([[ 2.7548, -1.1361, -2.3317]])
linear.bias : tensor([0.5757])
## Loss at the end of training = 0.17764988541603088
3.3.3. Learning Rate Optimization#
Exercise
Work with polynomial degree = 3.
Apply grid and/or random search over the learning rate of Adam. Do also combined optimization over the second order momentum parameter \(\beta_2\) and learning rate.
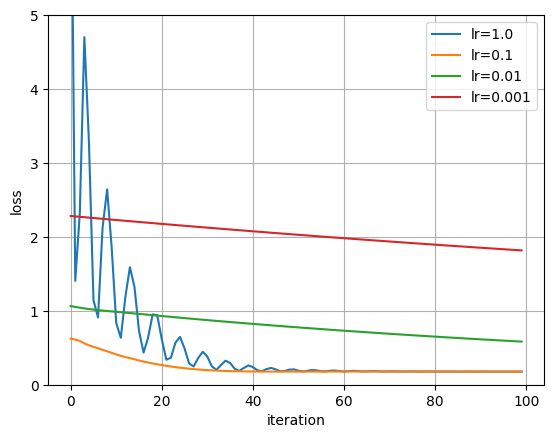
# TODO:
# grid search over learning rate for 100 steps
polynomial_degree = 3
num_iters = 100
lrs = [1.0, 0.1, 0.01, 0.001]
fig = plt.figure()
for lr in lrs:
model = PolynomialModel(polynomial_degree)
# Use LBFGS for fast convergence, but check validity of solution.
optimizer = torch.optim.Adam(model.parameters(), lr=lr)
criterion = nn.MSELoss()
losses = []
for iter in range(num_iters):
def closure():
optimizer.zero_grad()
y_pred = model(x_data)
loss = criterion(y_pred, y_data)
loss.backward()
return loss
optimizer.step(closure)
losses.append(closure().detach().item())
plt.plot(losses, label=f"lr={lr}")
plt.xlabel("iteration")
plt.ylabel("loss")
plt.legend()
plt.grid()
plt.ylim((0, 5))
(0.0, 5.0)
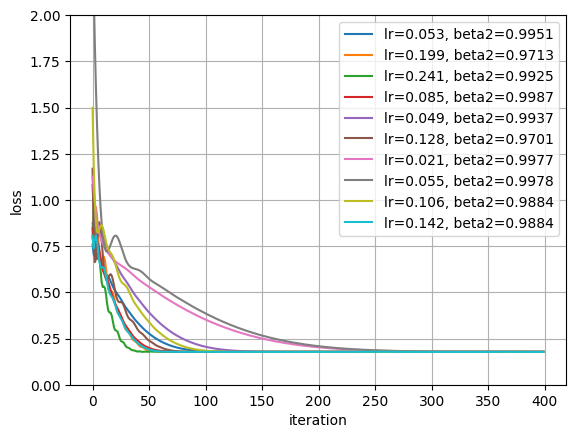
# random search over learning rate and beta_2 for 1k steps
def sample_loguniform(lb, ub, n):
"""Sapmle from the log-uniform distribution.
This is very useful for explore different orders of magnitude.
See e.g. this article: https://towardsdatascience.com/why-is-the-log-uniform-distribution-useful-for-hyperparameter-tuning-63c8d331698
lb: lower boundary
ub: upper boundary
n: number of samples
"""
return np.exp(np.random.uniform(np.log(lb), np.log(ub), n,))
polynomial_degree = 3
num_iters = 400
np.random.seed(123)
torch.manual_seed(123)
# log-uniform samples on the interval (0.5-0.02)
lrs = sample_loguniform(0.5, 0.02, 10)
# log-uniform samples on the interval (0.9, 0.999) finer resolved towards 1
beta2s = 1 - sample_loguniform(0.001, 0.1, 10)
# now train the model at each of these points
fig = plt.figure()
final_losses = []
for (lr, beta2) in zip(lrs, beta2s):
model = PolynomialModel(polynomial_degree)
# Use LBFGS for fast convergence, but check validity of solution.
optimizer = torch.optim.Adam(model.parameters(), lr=lr, betas=(0.9, beta2))
criterion = nn.MSELoss()
losses = []
for iter in range(num_iters):
def closure():
optimizer.zero_grad()
y_pred = model(x_data)
loss = criterion(y_pred, y_data)
loss.backward()
return loss
optimizer.step(closure)
losses.append(closure().detach().item())
final_losses.append(losses[-1])
plt.plot(losses, label=f"lr={lr:0.3f}, beta2={beta2:.4f}")
plt.xlabel("iteration")
plt.ylabel("loss")
plt.legend()
plt.ylim((0, 2))
plt.grid()
plt.show()
# second scatter plot
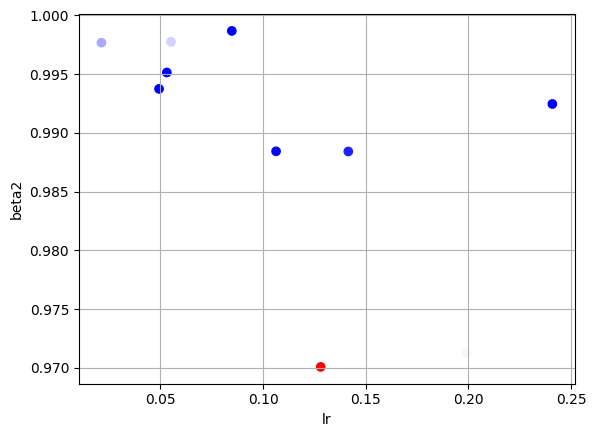
fig2 = plt.figure()
final_losses = np.array(final_losses)
plt.scatter(lrs, beta2s, c=final_losses, cmap='bwr')
plt.xlabel("lr")
plt.ylabel("beta2")
plt.grid()
idx = final_losses == final_losses.min()
print(f"Our best guess is lr={lrs[idx]}, beta2={beta2s[idx]}, and loss = {final_losses[idx]}")
Our best guess is lr=[0.2409047], beta2=[0.99246394], and loss = [0.17758173]
3.3.4. Learning Rate Scheduling#
Exercise
Work with polynomial degree = 3.
Apply stepwise decreasing learning rate - in PyTorch MultiStepLR using the Adam optimizer.
from torch.optim.lr_scheduler import MultiStepLR
# TODO:
# MultiStepLR implements a learning rate which descreeses by a factor of
# `gamma` at each of the given `milestones`, i.e. steps during optimization.
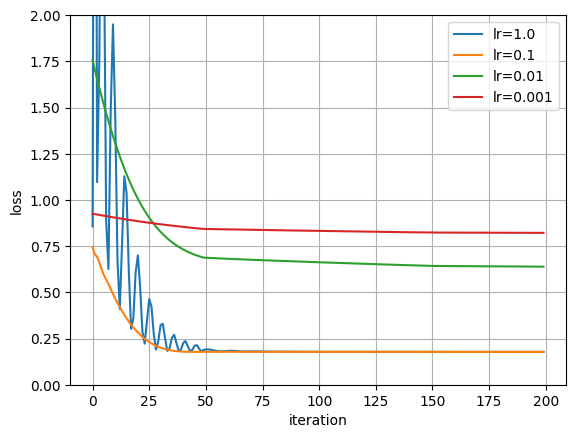
polynomial_degree = 3
num_iters = 200
lr_inits = [1.0, 0.1, 0.01, 0.001]
fig = plt.figure()
for lr in lr_inits:
model = PolynomialModel(polynomial_degree)
# This learning rate `lr` given to the optimizer is the initial one
optimizer = torch.optim.Adam(model.parameters(), lr=lr)
scheduler = MultiStepLR(optimizer, milestones=[50, 150], gamma=0.2)
criterion = nn.MSELoss()
losses = []
for iter in range(num_iters):
def closure():
optimizer.zero_grad()
y_pred = model(x_data)
loss = criterion(y_pred, y_data)
loss.backward()
return loss
optimizer.step()
scheduler.step()
losses.append(closure().detach().item())
plt.plot(losses, label=f"lr={lr}")
plt.xlabel("iteration")
plt.ylabel("loss")
plt.legend()
plt.grid()
plt.ylim((0, 2))
plt.show()
# In this plot it is not apparent why LR Scheduler are so necessary, but you
# will see the true benefit when we start working with deep learning models.
3.3.5. Regularization#
Exercise
Work with polynomial degree = 5.
Apply L1 and L2 regularization to the weights of the model using the Adam optimizer.
Hint: See this discussion. More precisely,
l1_norm = sum(torch.linalg.norm(p, 1) for p in model.parameters())andl2_norm = sum(torch.linalg.norm(p, 2) for p in model.parameters())
# TODO:
# # Parameters
torch.manual_seed(42)
np.random.seed(42)
num_iters = 10000
# weighting value for regularization term in the loss
reg_lambda = 0.1
dataset_case = "default" # "default", "polynomial" or "multivariate"
if dataset_case == "default":
polynomial_degree = 5
elif dataset_case == "polynomial":
np.random.seed(42)
n_samples = 20
x_data = torch.rand(size=(n_samples, 1)) * 3 - 1.5 # Uniformly distributed between -1.5 and 1.5
y_data = x_data**2 + 0.01 * torch.randn_like(x_data)
polynomial_degree = 10
elif dataset_case == "multivariate":
n_samples = 20
num_features = 10 # number of features
coeff_target = torch.zeros(num_features)
coeff_target[:3] = 1.0
x_data = torch.randn(n_samples, num_features)
y_data = x_data @ coeff_target + 0.1 * torch.randn(n_samples)
y_data = y_data.unsqueeze(1)
def no_reg(model):
"""no regularization"""
return 0
def l1_reg(model):
"""l1 regularization"""
return sum(torch.norm(p, 1) for p in model.parameters())
def l2_reg(model):
"""l2 regularization
Using `torch.optim.Adam`, this regularization is already there under the
parameter `weight_decay`
"""
return sum(torch.norm(p, 2) for p in model.parameters())
for case, reg in zip(["no", "L1", "L2"], [no_reg, l1_reg, l2_reg]):
print(f"################# Training with {case} regularization ###################")
losses = []
if dataset_case in ["polynomial", "default"]:
model = PolynomialModel(polynomial_degree)
elif dataset_case == "multivariate":
model = nn.Linear(num_features, 1)
# optimizer = torch.optim.SGD(model.parameters(), lr=0.003)
optimizer = torch.optim.Adam(model.parameters(), lr=0.01)
# optimizer = torch.optim.LBFGS(model.parameters())
criterion = nn.MSELoss()
print("## Parameters before training")
for name, param in model.named_parameters():
print(name, ": ", param.data)
for iter in range(num_iters):
def closure():
optimizer.zero_grad()
y_pred = model(x_data)
# here we apply regularization
loss = criterion(y_pred, y_data) + reg_lambda * reg(model)
loss.backward()
return loss
optimizer.step(closure)
losses.append(closure().detach().item())
print("## Parameters after training:")
torch.set_printoptions(sci_mode=False)
for name, param in model.named_parameters():
print(name, ": ", param.data)
if dataset_case in ["polynomial", "default"]:
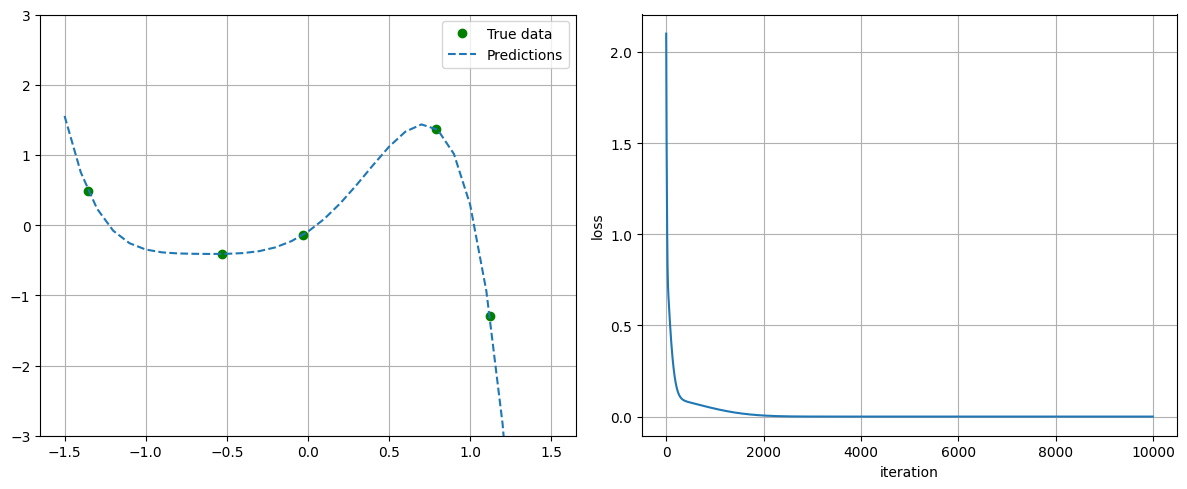
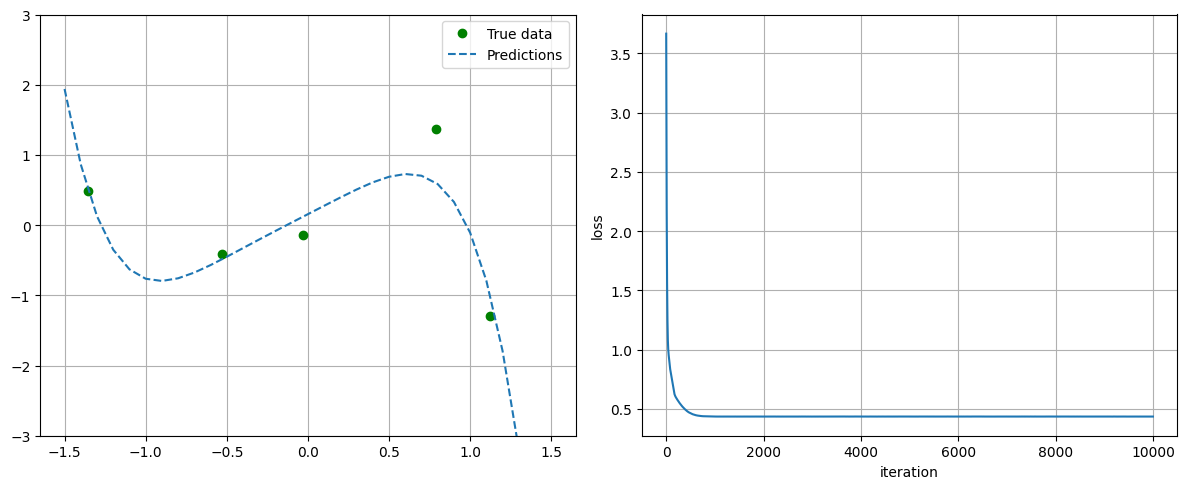
plot_progress_polynomial(losses, model, x_data, y_data)
################# Training with no regularization ###################
## Parameters before training
linear.weight : tensor([[ 0.3419, 0.3712, -0.1048, 0.4108, -0.0980]])
linear.bias : tensor([0.0902])
## Parameters after training:
linear.weight : tensor([[ 1.5655, 2.3635, 0.2018, -2.2983, -1.4480]])
linear.bias : tensor([-0.0935])
################# Training with L1 regularization ###################
## Parameters before training
linear.weight : tensor([[-0.2177, 0.2626, 0.3942, -0.3281, 0.3887]])
linear.bias : tensor([0.0837])
## Parameters after training:
linear.weight : tensor([[ 1.1930, 0.0004, 0.0004, -0.5924, -0.8655]])
linear.bias : tensor([0.1560])
################# Training with L2 regularization ###################
## Parameters before training
linear.weight : tensor([[ 0.3304, 0.0606, 0.2156, -0.0631, 0.3448]])
linear.bias : tensor([0.0661])
## Parameters after training:
linear.weight : tensor([[ 1.3985, 0.7845, 0.0293, -1.1680, -1.0786]])
linear.bias : tensor([0.0590])